PRT802: Difference between revisions
From ActinoBase
Sam Prudence (talk | contribs) |
Sam Prudence (talk | contribs) No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
==Use== | ==Use== | ||
Integrative vector is for general use in ''Streptomyces''. For example, producing marked strains for re-isolation experiments, or gene expression, over expression, or complementation. This vector can be replicated within ''Escherichia coli'' for cloning, upon transformation into ''Streptomyces'' via [[Tri-parental mating]] this vector will integrate into the genome at the ΦBT1 site. | Integrative vector is for general use in ''Streptomyces''. For example, producing marked strains for re-isolation experiments, or gene expression, over expression, or complementation. This vector can be replicated within ''Escherichia coli'' for cloning, upon transformation into ''Streptomyces'' via [[Tri-parental mating]] this vector will integrate into the genome at the ΦBT1 ''attB'' site. | ||
==Features== | ==Features== | ||
*Kanamycin resistance gene | *Kanamycin resistance gene | ||
*ΦBT1 phage integration site and integrase | *ΦBT1 ''attB'' phage integration site and integrase | ||
*Multiple cloning site | *Multiple cloning site | ||
*Origin of conjugative transfer (''oriT'') | *Origin of conjugative transfer (''oriT'') | ||
| Line 18: | Line 18: | ||
==Sequence links== | ==Sequence links== | ||
[https://www.addgene.org/68113/ addgene page] | See the [https://www.addgene.org/68113/ addgene page], click "sequence" to download .gbk file. | ||
==References== | ==References== | ||
# Gregory, M.A., Till, R., Smith, M.C.M. (2003). Integration Site for Streptomyces Phage ΦBT1 and Development of | # Gregory, M.A., Till, R., Smith, M.C.M. (2003). Integration Site for Streptomyces Phage ΦBT1 and Development of Site-Specific Integrating Vectors. ''Journal of Bacteriology'', 185(17), pp. 5320-5323. DOI: 10.1128/JB.185.17.5320–5323.2003 | ||
Site-Specific Integrating Vectors. ''Journal of Bacteriology'', 185(17), pp. 5320-5323. DOI: 10.1128/JB.185.17.5320–5323.2003 | |||
Latest revision as of 10:08, 9 August 2019
Use
Integrative vector is for general use in Streptomyces. For example, producing marked strains for re-isolation experiments, or gene expression, over expression, or complementation. This vector can be replicated within Escherichia coli for cloning, upon transformation into Streptomyces via Tri-parental mating this vector will integrate into the genome at the ΦBT1 attB site.
Features
- Kanamycin resistance gene
- ΦBT1 attB phage integration site and integrase
- Multiple cloning site
- Origin of conjugative transfer (oriT)
- Origin of replication for E. coli (pBR322oriF)
History
Backbone vector pSET152 was modified to replace the ΦC31 integrative machinery with the ΦBT1 integrative machinery by Maggie Smiths group, published in 20031.
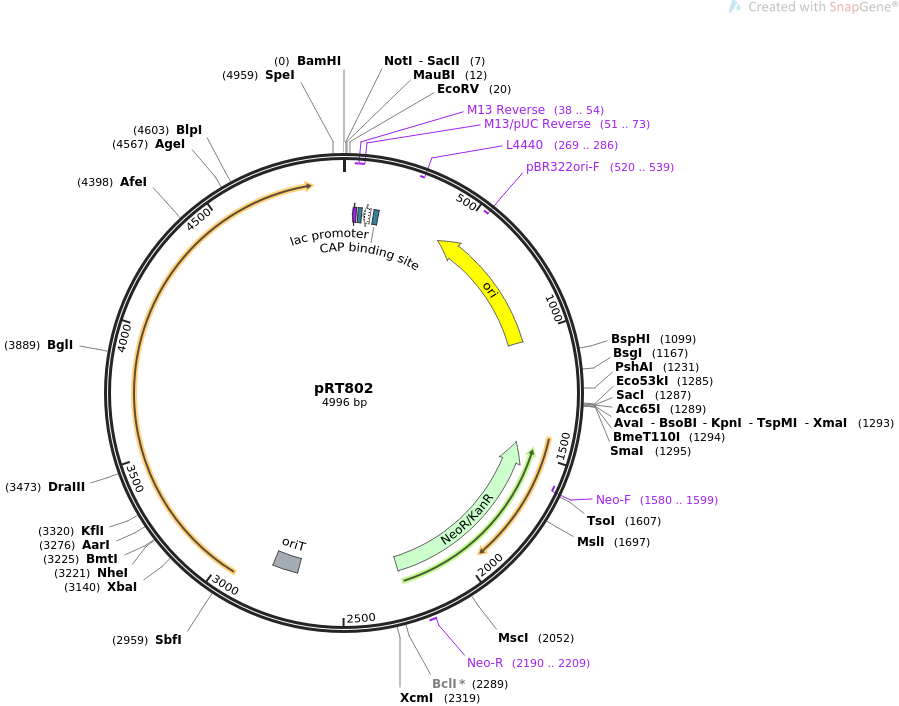
Map
Sequence links
See the addgene page, click "sequence" to download .gbk file.
References
- Gregory, M.A., Till, R., Smith, M.C.M. (2003). Integration Site for Streptomyces Phage ΦBT1 and Development of Site-Specific Integrating Vectors. Journal of Bacteriology, 185(17), pp. 5320-5323. DOI: 10.1128/JB.185.17.5320–5323.2003