PCRISPomyces-2: Difference between revisions
From ActinoBase
Sam Prudence (talk | contribs) No edit summary |
Sam Prudence (talk | contribs) No edit summary |
||
| Line 2: | Line 2: | ||
==Use== | |||
This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of <em>Streptomyces</em> strains. To our knowledge it has been successfully used in [https://doi.org/10.1128/mSphere.00305-16 S. albidoflavus], S. coelicolor, [https://pubs.rsc.org/en/content/articlelanding/2017/SC/C6SC04265A#!divAbstract S. formicae], [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4459934/ S. lividans] and S. venezuelae. Mutations have ranged from 1-100kbp deletions, precise codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci. | This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of <em>Streptomyces</em> strains. To our knowledge it has been successfully used in [https://doi.org/10.1128/mSphere.00305-16 S. albidoflavus], S. coelicolor, [https://pubs.rsc.org/en/content/articlelanding/2017/SC/C6SC04265A#!divAbstract S. formicae], [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4459934/ S. lividans] and S. venezuelae. Mutations have ranged from 1-100kbp deletions, precise codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci. | ||
| Line 9: | Line 9: | ||
Click here for the [[CRISPR]] protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - [http://www.hutchingslab.uk/downloads/pCRISPOmyces2.pdf or download the PDF]. | Click here for the [[CRISPR]] protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - [http://www.hutchingslab.uk/downloads/pCRISPOmyces2.pdf or download the PDF]. | ||
==Features== | |||
==History== | |||
The plasmid was made by [http://faculty.scs.illinois.edu/~zhaogrp/index.html Huimin Zhao's] group and is available free from AddGene under MTA: [https://www.addgene.org/61737/ click here] | The plasmid was made by [http://faculty.scs.illinois.edu/~zhaogrp/index.html Huimin Zhao's] group and is available free from AddGene under MTA: [https://www.addgene.org/61737/ click here] | ||
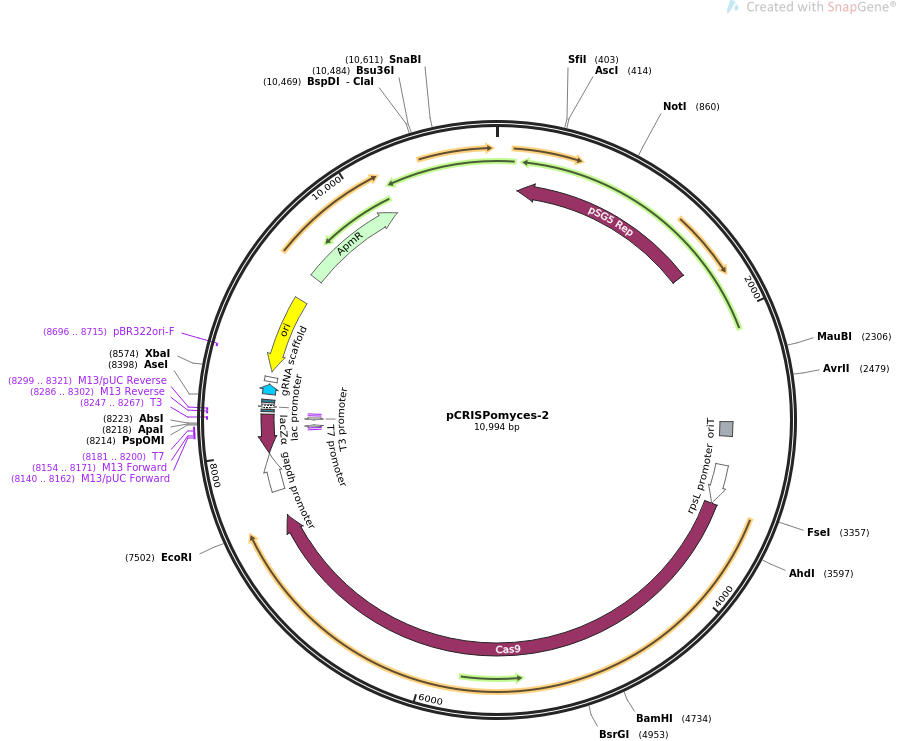
[[file: | ==Map== | ||
[[file:PCRISPomyces-2_map.png]] | |||
==Sequence links== | |||
AddGene under MTA: [https://www.addgene.org/61737/ click here] | |||
==References== | |||
Revision as of 16:53, 8 August 2019
Use
This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of Streptomyces strains. To our knowledge it has been successfully used in S. albidoflavus, S. coelicolor, S. formicae, S. lividans and S. venezuelae. Mutations have ranged from 1-100kbp deletions, precise codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci.
Click here for the CRISPR protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - or download the PDF.
Features
History
The plasmid was made by Huimin Zhao's group and is available free from AddGene under MTA: click here
Map
Sequence links
AddGene under MTA: click here