PCRISPomyces-2: Difference between revisions
From ActinoBase
Sam Prudence (talk | contribs) No edit summary |
Sam Prudence (talk | contribs) |
||
| Line 10: | Line 10: | ||
==Features== | ==Features== | ||
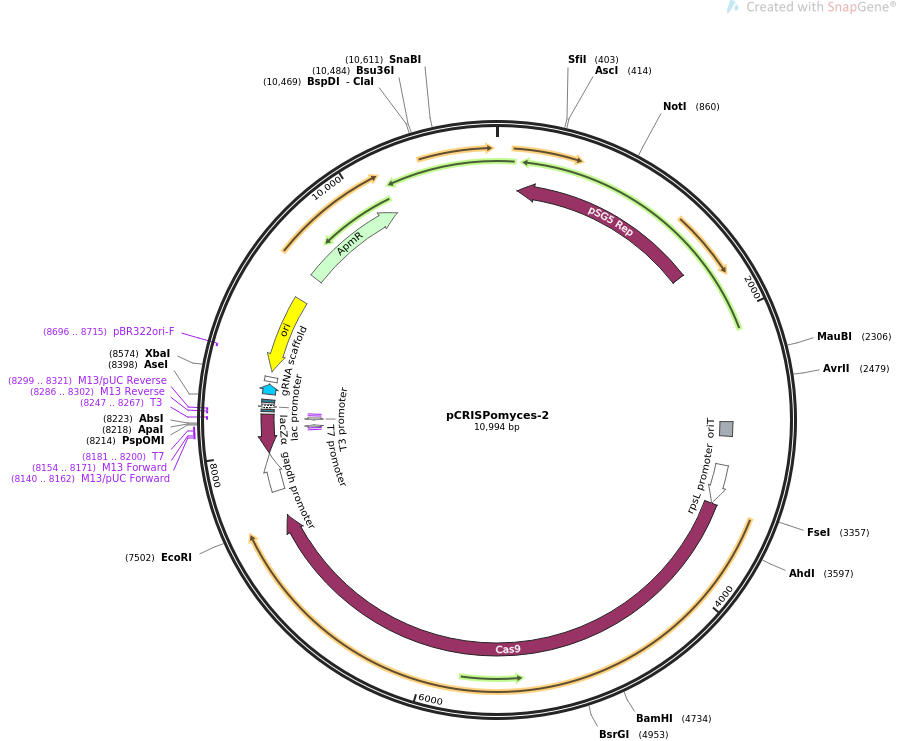
*Ampicillin resistance cassette | |||
*Origin of conjugative transfer (''oriT'') | |||
*Origin of replication for ''Escherichia coli'' (pBR322''oriF'') | |||
*Codon optomised ''cas9'' gene under the constitutive rpSL promotor | |||
====Unfinished==== | |||
==History== | ==History== | ||
Revision as of 09:36, 9 August 2019
Use
This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of Streptomyces strains. To our knowledge it has been successfully used in S. albidoflavus, S. coelicolor, S. formicae, S. lividans and S. venezuelae. Mutations have ranged from 1-100kbp deletions, precise codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci.
Click here for the CRISPR protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - or download the PDF.
Features
- Ampicillin resistance cassette
- Origin of conjugative transfer (oriT)
- Origin of replication for Escherichia coli (pBR322oriF)
- Codon optomised cas9 gene under the constitutive rpSL promotor
Unfinished
History
The plasmid was made by Huimin Zhao's group and is available free from AddGene under MTA: click here
Map
Sequence links
AddGene under MTA: click here