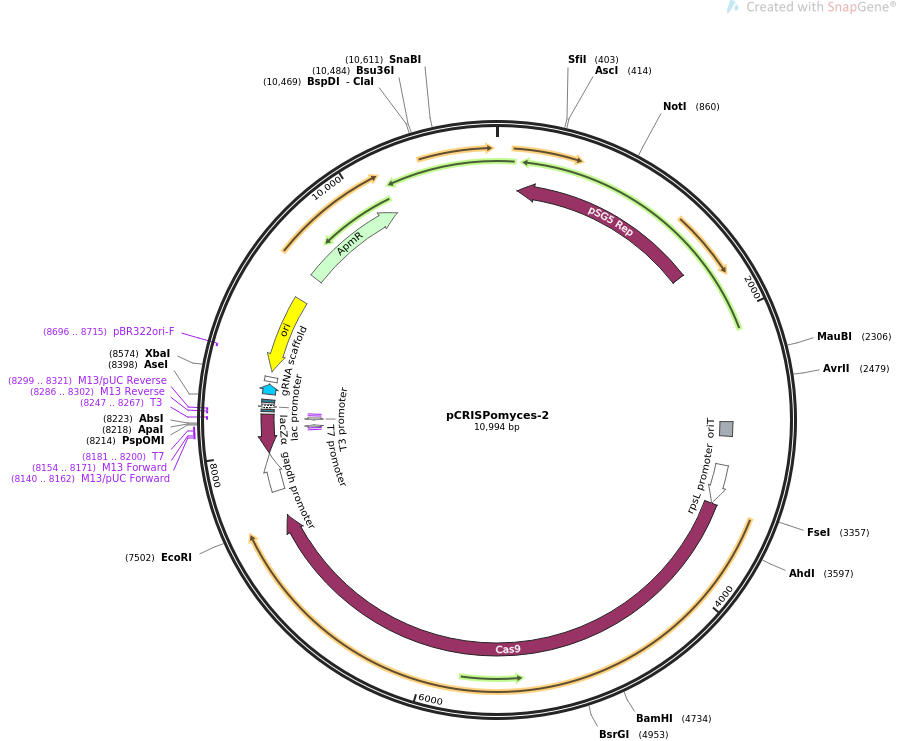
PCRISPomyces-2
From ActinoBase
Use
This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of Streptomyces strains. To our knowledge it has been successfully used in S. albidoflavus, S. coelicolor, S. formicae, S. lividans and S. venezuelae. Mutations have ranged from 1-100kbp deletions, precise codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci.
Click here for the CRISPR protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - or download the PDF.
Features
- Ampicillin resistance cassette
- Origin of conjugative transfer (oriT)
- Origin of replication for Escherichia coli (pBR322oriF)
- Codon optomised cas9 gene under the constitutive rpSL promoter
- LacZ cassette for golden gate cloning of guide RNA (gRNA)
- Constitutive gapdh promoter for gRNA expression
- pSG5 temparature sensitive Streptomyces origin of replication. Without selection and above 37°C the plasmid becomes unstable. This property is used to cure the plasmid from Streptomyces after successful mutagenesis.
- Xba1 site for gibson assembly of repair templates
History
The plasmid was made by Huimin Zhao's group and is available free from AddGene under MTA: click here
Map
Sequence links
AddGene under MTA: click here