RRNA depletion with Streptomyces coelicolor A3(2) specific probes: Difference between revisions
(→Notes) |
(→Notes) |
||
| Line 25: | Line 25: | ||
==Notes== | ==Notes== | ||
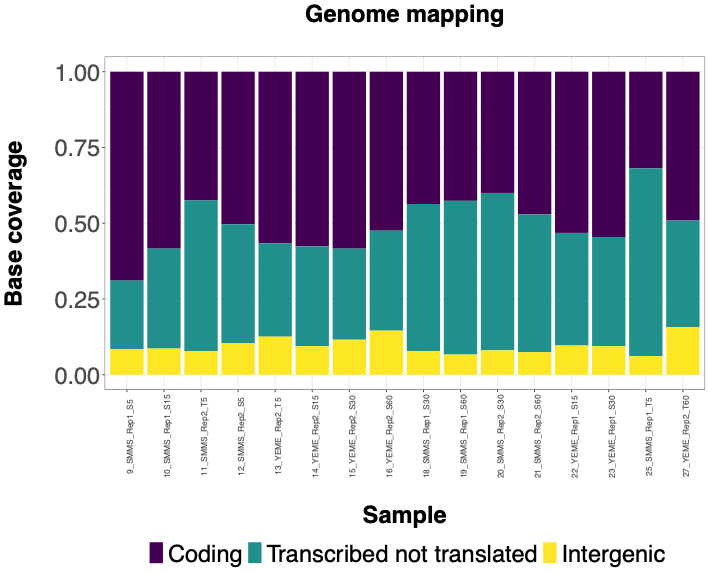
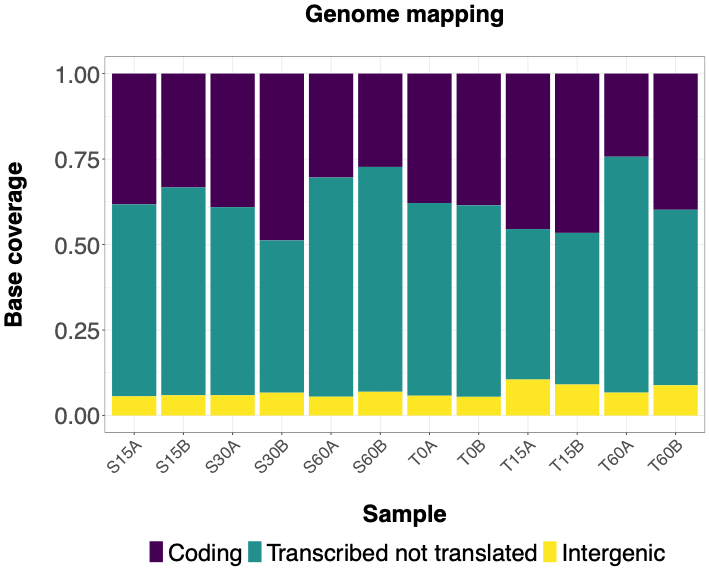
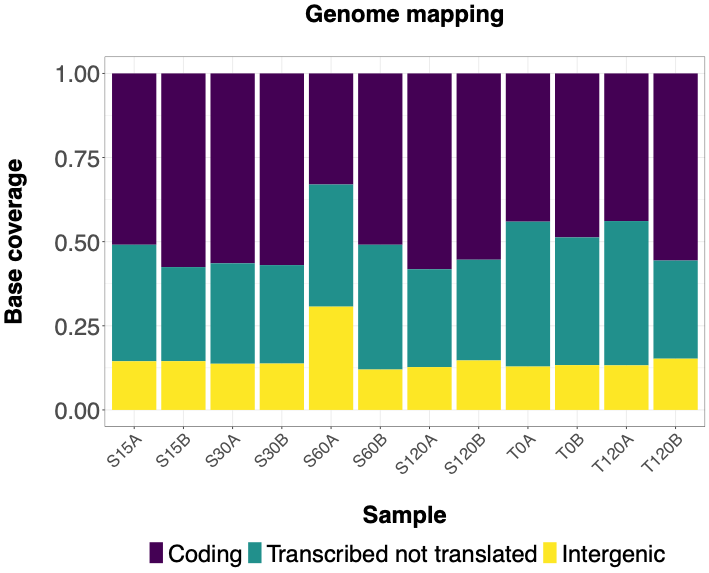
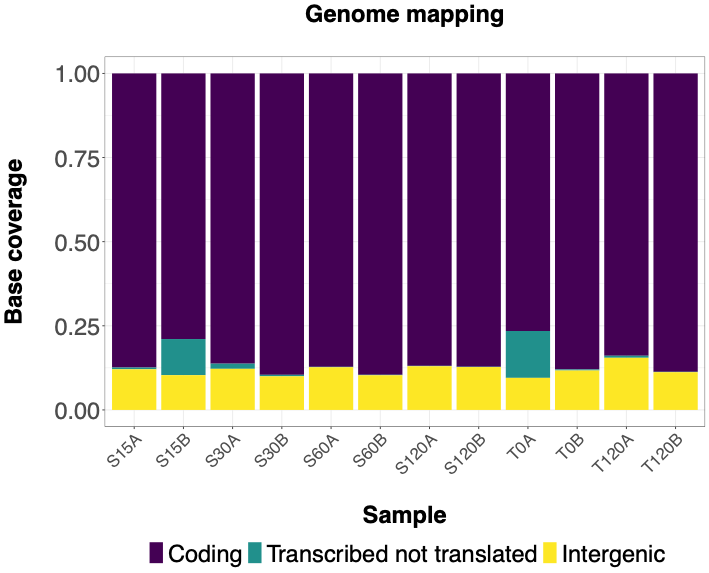
The following histograms illustrate effectiveness of rRNA depletion using various methods. Coding = RNA aligned to coding sequences; Transcribed not translated = RNA aligned to rRNA, tRNA and sRNA; Intergenic = RNA aligned to 5' or 3' UTRs or between annotated sequences. | The following histograms illustrate effectiveness of rRNA depletion using various methods. Histograms show percentage of bases aligned to NCBI reference sequence NC_003888.3 using Hisat2 program classified as follows - Coding = RNA aligned to coding sequences; Transcribed not translated = RNA aligned to rRNA, tRNA and sRNA; Intergenic = RNA aligned to 5' or 3' UTRs or between annotated sequences. | ||
'''QIAseq FastSelect 5S/16S/23S kit''' | '''QIAseq FastSelect 5S/16S/23S kit''' | ||
Revision as of 16:01, 1 November 2023
Organisms
This protocol has been tested with Streptomyces coelicolor A3(2) MT1110
Description
rRNA depletion using commercial kits targeting generic bacterial sequences is not 100% efficient in the High GC organism S. coelicolor.
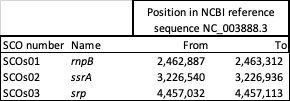
Depletion using the NEBNext RNA Depletion Core Reagent Set with RNA Sample Purification Beads (https://www.neb.com/en-gb/products/e7870-nebnext-rna-depletion-core-reagent-set-with-rna-sample-purification-beads#Product%20Information) and custom DNA probes designed for S. coelicolor is more effective than the QIAseq FastSelect 5S/16S/23S kit (Qiagen #335927); NEBNext rRNA Depletion Kit (Bacteria) (NEB #E7850); Illumina Ribo-zero rRNA Removal Kit (Bacteria) (Illumina #MRZMB12424, discontinued). The Illumina kit tested was subsequently replaced by the Illumina Ribo-Zero Plus rRNA Depletion Kit (Illumina #20040526). Additionally three abundant non-mRNA RNAs can be depleted (Table 1).
Table 1. Abundant non-mRNA RNAs in S. coelicolor.
Materials
Single stranded DNA probes (Table 2) at 2µM concentration in IDTE pH 7.5 (10mM Tris-HCl / 0.1mM EDTA). Integrated DNA Technologies can produce these from their 25nmole DNA oligo pool product.
NEBNext RNA Depletion Core Reagent Set with RNA Sample Purification Beads (https://www.neb.com/en-gb/products/e7870-nebnext-rna-depletion-core-reagent-set-with-rna-sample-purification-beads#Product%20Information).
Protocols
As per manufacturer’s instructions.
Notes
The following histograms illustrate effectiveness of rRNA depletion using various methods. Histograms show percentage of bases aligned to NCBI reference sequence NC_003888.3 using Hisat2 program classified as follows - Coding = RNA aligned to coding sequences; Transcribed not translated = RNA aligned to rRNA, tRNA and sRNA; Intergenic = RNA aligned to 5' or 3' UTRs or between annotated sequences.
QIAseq FastSelect 5S/16S/23S kit
NEBNext rRNA Depletion Kit (Bacteria)
Illumina Ribo-zero rRNA Removal Kit (Bacteria)
rRNA depletion with S. coelicolor specific probes