RRNA depletion with Streptomyces coelicolor A3(2) specific probes
Organisms
This protocol has been tested with Streptomyces coelicolor A3(2) MT1110
rRNA depletion with Streptomyces coelicolor A3(2) specific probes
rRNA depletion using commercial kits targeting generic bacterial sequences is not 100% efficient in the High GC organism S. coelicolor.
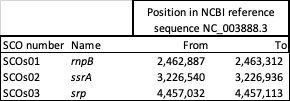
Depletion using the NEBNext RNA Depletion Core Reagent Set with RNA Sample Purification Beads (https://www.neb.com/en-gb/products/e7870-nebnext-rna-depletion-core-reagent-set-with-rna-sample-purification-beads#Product%20Information) and custom DNA probes designed for S. coelicolor is more effective than the QIAseq FastSelect 5S/16S/23S kit (Qiagen #335927); NEBNext rRNA Depletion Kit (Bacteria) (NEB #E7850); Illumina Ribo-zero rRNA Removal Kit (Bacteria) (Illumina #MRZMB12424, discontinued). The Illumina kit tested was subsequently replaced by the Illumina Ribo-Zero Plus rRNA Depletion Kit (Illumina #20040526). Additionally three abundant non-mRNA RNAs can be depleted (Table 1).