PCRISPomyces-2: Difference between revisions
From ActinoBase
No edit summary |
Sam Prudence (talk | contribs) |
||
| (23 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE: pCRISPomyces-2}} | |||
==Use== | |||
This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of <em>Streptomyces</em> strains</em><sup>1</sup>. It is possible to introduce mutations that range from 1-100kbp deletions, precise single base codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci. | |||
This plasmid has been successfully used for genome editing in the following organisms | |||
*<em>[[Actinoplanes]]</em> sp. SE50/110</em><sup>2</sup> | |||
*<em>[[Streptomyces albidoflavus]]</em><sup>3</sup> | |||
*<em>[[Streptomyces coelicolor]]</em> (unpulished mutagenesis performed within the group of [https://people.uea.ac.uk/m_hutchings Prof Matt Hutchings]) | |||
*<em>[[Streptomyces formicae]]</em><sup>4</sup> | |||
*<em>[[Streptomyces lividans]]</em><sup>1</sup> | |||
*<em>[[Streptomyces rimosus]]</em></em></em><sup>5</sup> | |||
*<em>[[Streptomyces roseosporus]]</em></em><sup>6</sup> | |||
*<em>[[Streptomyces showdoensis]]</em><sup>7</sup> | |||
*<em>[[Streptomyces venezuelae]]</em></em><sup>6</sup> | |||
*<em>[[Streptomyces viridochromogenes]]</em></em><sup>1</sup> | |||
[[file: | |||
See here for the original [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4459934/ pCRISPomyces-2 paper] from Cobb ''et. al''</em><sup>1</sup>. | |||
Click here for the [[CRISPR]] protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - [http://www.hutchingslab.uk/downloads/pCRISPOmyces2.pdf or download the PDF]. | |||
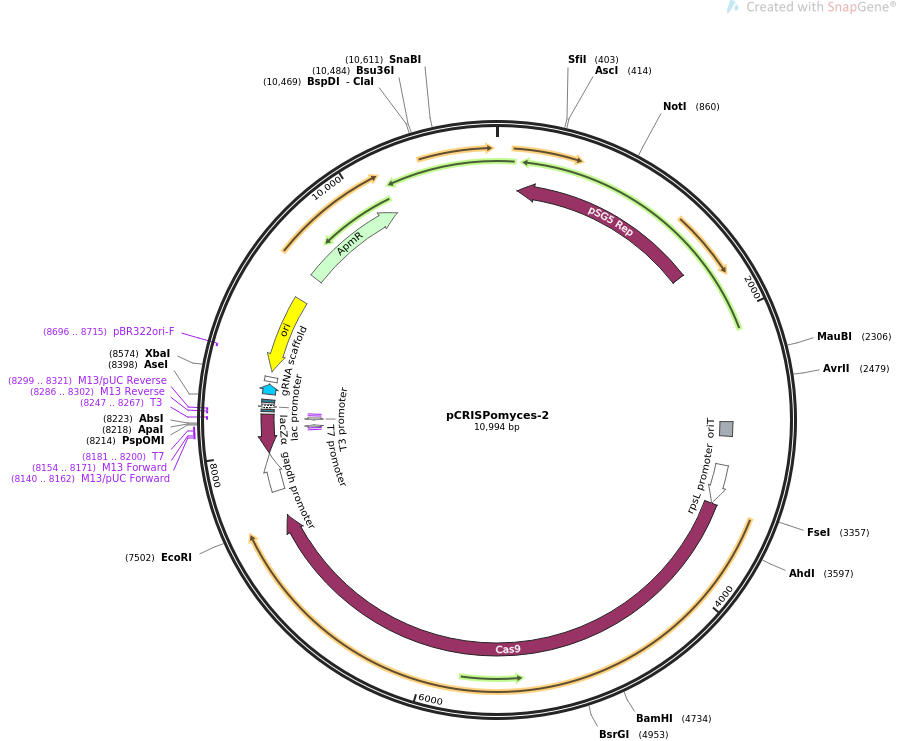
==Features== | |||
*Apramycin resistance cassette | |||
*Origin of conjugative transfer (''oriT'') | |||
*Origin of replication for ''Escherichia coli'' (pBR322''oriF'') | |||
*Codon optomised ''cas9'' gene under the constitutive ''rpSL'' promoter | |||
*LacZ cassette for golden gate cloning of guide RNA (gRNA) | |||
*Constitutive ''gapdh'' promoter for gRNA expression | |||
*''pSG5'' temparature sensitive ''Streptomyces'' origin of replication. Without selection and above 37°C the plasmid becomes unstable. This property is used to cure the plasmid from ''Streptomyces'' after successful mutagenesis. | |||
*Xba1 site for gibson assembly of repair templates | |||
==History== | |||
The plasmid was made by [http://faculty.scs.illinois.edu/~zhaogrp/index.html Huimin Zhao's] group and is available free from AddGene under MTA: [https://www.addgene.org/61737/ click here] | |||
==Map== | |||
[[file:PCRISPomyces-2_map.png]] | |||
==Sequence links== | |||
AddGene under MTA: [https://www.addgene.org/61737/ click here] | |||
==References== | |||
#Cobb R.E., Wang, Y., Zhao, H. (2014). High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ''ACS Synthetic Biology'', 4(6), pp. 723-728. DOI: 10.1021/sb500351f | |||
#Wolf, T., Gren, T., Thieme, E., Wibberg, D., Zemke, T., Pühler, A., Kalinowski, J. (2016). Targeted genome editing in the rare actinomycete Actinoplanes sp. SE50/110 by using the CRISPR/Cas9 System. ''Journal of Biotechnology'', 231, pp. 122-128. DOI: 10.1016/j.jbiotec.2016.05.039. | |||
#McLean, T.C., Hoskisson, P.A., Seipke, R.F. (2016). Coordinate Regulation of Antimycin and Candicidin Biosynthesis. ''mSphere'', 1(6), pp. e00305-e00316. DOI: 10.1128/mSphere.00305-16. | |||
#Qin, Z., Munnoch, J.T., Devine R., Holmes, N.A., Seipke, R.F., Wilkinson, K.A., Wilkinson., Hutchings, M.H. (2017). Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. ''Chemical Science'', 8, pp. 3218-3227. DOI: 10.1039/c6sc04265aT | |||
#Jia, H., Zhang, L., Wang, T., Han, J., Tang, H., Zhang, L. (2017). Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus. ''Microbiology'', 163(8), pp. 1148-1155. DOI: 10.1099/mic.0.000501. | |||
#Zhang, M.M., Wong, F.T., Wang, Y., Luo, S., Lim, Y.H., Heng, H., Yeo, W.L., Cobb, R.E., Enghiad, B., Ang, E.L., Zhao, H. (2017). Crispr–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. ''Nature Chemical Biology'', 13, pp. 607-609. DOI: 10.1038/nchembio.2341 | |||
#Palmu, K., Rosenqvist, P., Thapa, K., Illina, Y., Siitonen, V., Baral, B., Mäkinen, J., Belogurov., Virta, P., Niemi, J., Metsä-Ketelä, M. (2017). Discovery of the Showdomycin Gene Cluster from Streptomyces showdoensis ATCC 15227 Yields Insight into the Biosynthetic Logic of C-Nucleoside Antibiotics. ''ACS Chemical Biology'', 12(6), pp. 1472-1477. DOI: 10.1021/acschembio.7b00078. | |||
Latest revision as of 17:24, 7 November 2019
Use
This plasmid can be used for precise CRISPR/Cas9-mediated genome engineering of Streptomyces strains1. It is possible to introduce mutations that range from 1-100kbp deletions, precise single base codon changes to alter amino acids and insertions to add Flag-tags to proteins encoded at their native loci.
This plasmid has been successfully used for genome editing in the following organisms
- Actinoplanes sp. SE50/1102
- Streptomyces albidoflavus3
- Streptomyces coelicolor (unpulished mutagenesis performed within the group of Prof Matt Hutchings)
- Streptomyces formicae4
- Streptomyces lividans1
- Streptomyces rimosus5
- Streptomyces roseosporus6
- Streptomyces showdoensis7
- Streptomyces venezuelae6
- Streptomyces viridochromogenes1
See here for the original pCRISPomyces-2 paper from Cobb et. al1.
Click here for the CRISPR protocol used by Matt Hutchings lab (courtesy of Rebecca Devine) - or download the PDF.
Features
- Apramycin resistance cassette
- Origin of conjugative transfer (oriT)
- Origin of replication for Escherichia coli (pBR322oriF)
- Codon optomised cas9 gene under the constitutive rpSL promoter
- LacZ cassette for golden gate cloning of guide RNA (gRNA)
- Constitutive gapdh promoter for gRNA expression
- pSG5 temparature sensitive Streptomyces origin of replication. Without selection and above 37°C the plasmid becomes unstable. This property is used to cure the plasmid from Streptomyces after successful mutagenesis.
- Xba1 site for gibson assembly of repair templates
History
The plasmid was made by Huimin Zhao's group and is available free from AddGene under MTA: click here
Map
Sequence links
AddGene under MTA: click here
References
- Cobb R.E., Wang, Y., Zhao, H. (2014). High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ACS Synthetic Biology, 4(6), pp. 723-728. DOI: 10.1021/sb500351f
- Wolf, T., Gren, T., Thieme, E., Wibberg, D., Zemke, T., Pühler, A., Kalinowski, J. (2016). Targeted genome editing in the rare actinomycete Actinoplanes sp. SE50/110 by using the CRISPR/Cas9 System. Journal of Biotechnology, 231, pp. 122-128. DOI: 10.1016/j.jbiotec.2016.05.039.
- McLean, T.C., Hoskisson, P.A., Seipke, R.F. (2016). Coordinate Regulation of Antimycin and Candicidin Biosynthesis. mSphere, 1(6), pp. e00305-e00316. DOI: 10.1128/mSphere.00305-16.
- Qin, Z., Munnoch, J.T., Devine R., Holmes, N.A., Seipke, R.F., Wilkinson, K.A., Wilkinson., Hutchings, M.H. (2017). Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chemical Science, 8, pp. 3218-3227. DOI: 10.1039/c6sc04265aT
- Jia, H., Zhang, L., Wang, T., Han, J., Tang, H., Zhang, L. (2017). Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus. Microbiology, 163(8), pp. 1148-1155. DOI: 10.1099/mic.0.000501.
- Zhang, M.M., Wong, F.T., Wang, Y., Luo, S., Lim, Y.H., Heng, H., Yeo, W.L., Cobb, R.E., Enghiad, B., Ang, E.L., Zhao, H. (2017). Crispr–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nature Chemical Biology, 13, pp. 607-609. DOI: 10.1038/nchembio.2341
- Palmu, K., Rosenqvist, P., Thapa, K., Illina, Y., Siitonen, V., Baral, B., Mäkinen, J., Belogurov., Virta, P., Niemi, J., Metsä-Ketelä, M. (2017). Discovery of the Showdomycin Gene Cluster from Streptomyces showdoensis ATCC 15227 Yields Insight into the Biosynthetic Logic of C-Nucleoside Antibiotics. ACS Chemical Biology, 12(6), pp. 1472-1477. DOI: 10.1021/acschembio.7b00078.